Bacterial Diversity in Submarine Groundwater along the Coasts of the Yellow Sea
Submarine groundwater discharge (SGD) is one of the most significant pathways for the exchange of groundwater and/or source of nutrients, metals and carbon to the ocean, subsequently cause deleterious impacts on the coastal ecosystems. Microorganisms have been recognized as the important participators in the biogeochemical processes in the SGD. In this study, by utilizing 16S rRNA-based Illumina Miseq sequencing technology, we investigated bacterial diversity and distribution in both fresh well water and brackish recirculated porewater along the coasts in the Yellow Sea. The results showed that Actinobacteria and Betaproteobacteria, especially Comamonas spp. and Limnohabitans spp. were dominated in fresh well samples. Distinct patterns of bacterial communities were found among the porewater samples due to different locations, for examples, Cyanobacteria was the most abundant in the porewater samples far from the algal bloomed areas. The analysis of correlation between representative bacterial taxonomic groups and the contexture environmental parameters showed that fresh well water and brackish porewater might provide different nutrients to the coastal waters. Potential key bacterial groups such as Comamonas spp. may be excellent candidates for the bioremediation of the natural pollutants in the SGD. Our comprehensive understanding of bacterial diversity in the SGD along the coasts of the Yellow Sea will create a basis for designing the effective clean-up approach in-situ, and provide valuable information for the coastal management.
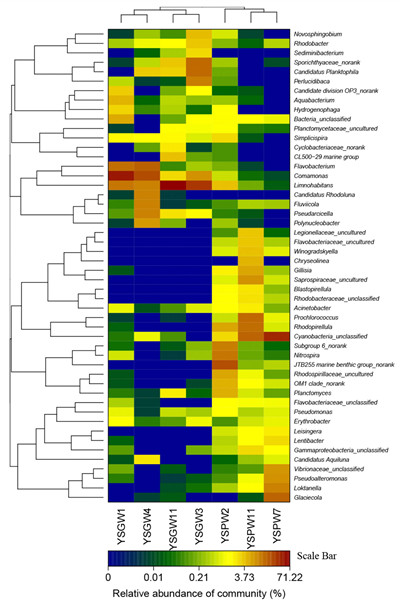
FIGURE2 Heatmap showing the relative abundance and distribution of genus-based OUT illumine reads. The color code indicates relative abundance, ranging from blue (low abundance) to black to brown (high abundance).
FIGURE7 Environmental factors associated with variations of the bacterial community structure at the phylum level. In order to obtain a higher resolution, the Proteobacteria phylum level was separated into Alpha, Beta, and Gamma-Proteobacteria classes. Phylum Bacteriodetes was separated into classes Flavabacterria and Cytophagia. The total number of sequences in each phylum is indicated in parentheses. Pearson’s correlation coefficients between-1 and 1 are shown in the rectangle, which indicates correlations between phylum/class sequence abundance and selected environmental parameters. For example, afirebrick colored rectangle (0.76) between nitrate and Betaproteobacteria indicates a higher number of sequences within creasing nitrate concentration, a blue rectangle (−0.774) between Betaproteobacteria and ammonium indicates a higher number of sequences with decreasing ammonium concentration. The color code indicates Pearson’s correlation coefficients, ranging from blue(−1) to white (0) to firebrick (1). The density showed the distribution of Pearson’s correlation coefficients between −1 and1.Si, silicate; P, phosphate; NO2, nitrite; NO3, nitrate; NH4, ammonium; 224Ra, Radium isotope tracer; DOC, dissolved organic carbon.
List of relate publications:
-
FRONTIERS IN MICROBIOLOGY ,
2016 ,
6
: 1519